Integrates with GitHub for accessing the OriGene project repository and other related resources for therapeutic research and development
Provides integration with LangChain through the MultiServerMCPClient, allowing connection to multiple biomedical tools and databases within the OrigeneMCP ecosystem
Enables using Python to interact with the OrigeneMCP toolkit through both direct API calls and the CAMEL framework for agent-based interactions
Supports configuration via TOML files for server settings, API keys, and other deployment parameters
OrigeneMCP
OrigeneMCP is a core component of the OriGene ecosystem, a self-evolving multi-agent system designed to act as a virtual disease biologist. OriGene provides a unified platform for biomedical AI research and was officially launched at the 2025 WAIC.
1. OrigeneMCP Overview
Recent updates:
2025-07-28 🚀 OriGene officially launched at WAIC 2025!
2025-07-22 🔥 Integrated OrigeneMCP into the Bohrium AI for Science platform.
2025-07-19 🔥 Integrated OrigeneMCP into CAMEL.
2025-07-18 🚀 OrigeneMCP officially open-sourced!
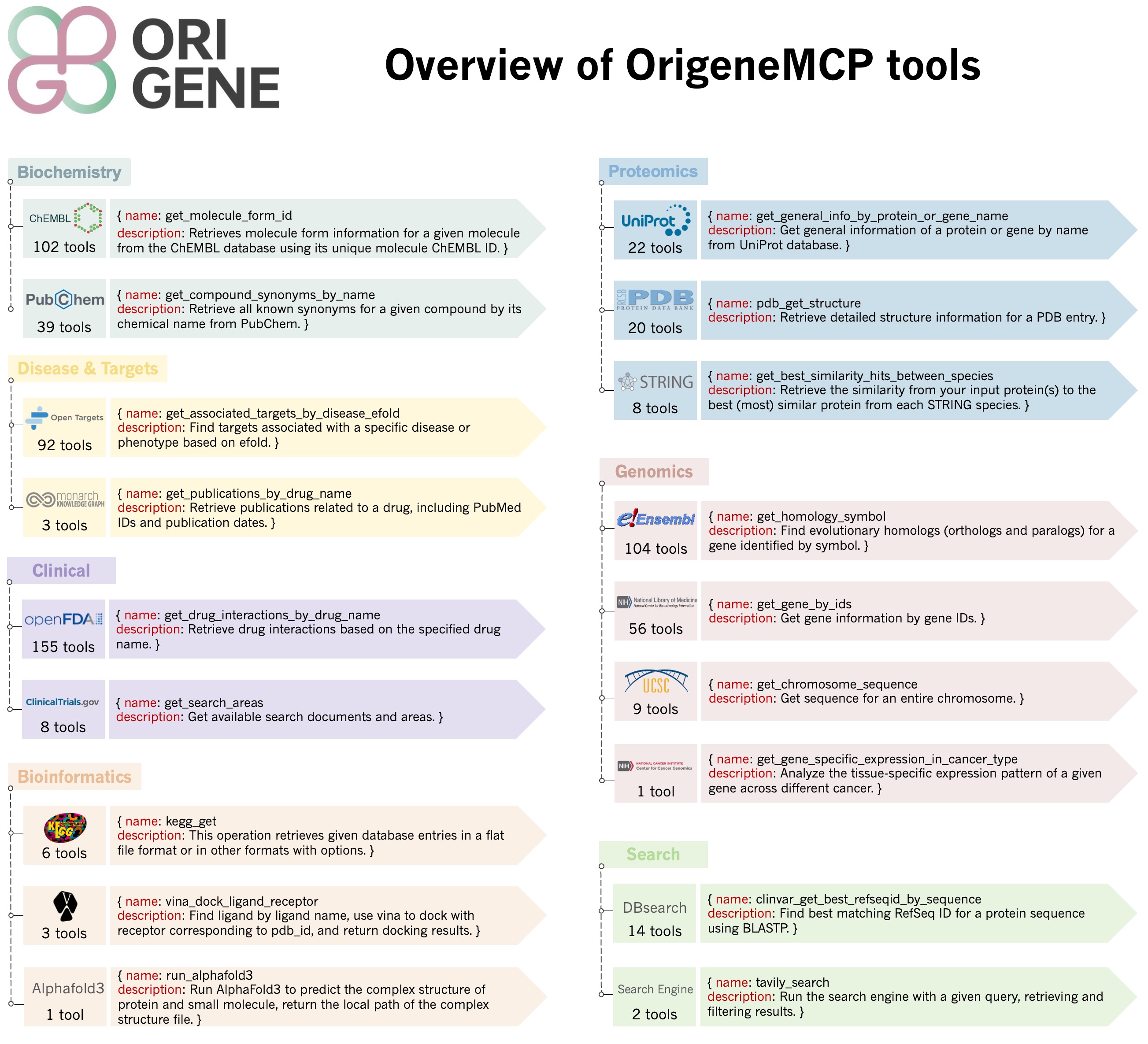
OrigeneMCP is the MCP toolkit for the OriGene project that integrates over 600 tools and multiple databases (including ChEMBL, PubChem, FDA, OpenTargets, NCBI, UniProt, PDB, Ensembl, UCSC, KEGG, STRING, TCGA, Monarch, ClinicalTrials, and more) into an advanced integrated MCP server platform, combining disease biology and drug discovery tools to enable comprehensive multi-dimensional information retrieval across small molecules, proteins, genes, diseases, and other biological entities, serving as a unified interface for accessing and analyzing complex biomedical data to accelerate therapeutic research and development.
Related MCP server: MCP Toolbox for Databases
2. Deploy OrigeneMCP
Clone the repository:
The dependencies of this repository are managed by uv. Before deployment, please make sure that uv is already installed. Use the following commands to install uv:
Initialize the dependencies of this repository:
Deploy the mcp service:
NOTE:
Please ensure that port 8788 is not occupied. If it is occupied, please modify the port in file
local.conf.tomlto another port;If you need to use Tavily search or Jina search, please configure the
tavily_api_keyorjina_api_keyin filelocal.conf.toml;
3. Use OrigeneMCP
3.1. Use locally deployed OrigeneMCP
First, connect to OrigeneMCP and get all available tools.
Let's explore a specific tool's capabilities by examining its description and test cases. As an example, we'll look at the get_general_info_by_protein_or_gene_name tool from UniProt, which provides comprehensive protein and gene information:
Set input parameters and call the tool:
3.2. Use OrigeneMCP through CAMEL
CAMEL is an open-source community dedicated to finding the scaling laws of agents. In addition to local invocation, we can also use OrigeneMCP-based Agents through the CAMEL interface.
Install the CAMEL package:
Set up your OpenAI API key:
Use the CAMEL interface to invoke the OrigeneMCP-based Agent:
3.3. Use OrigeneMCP through Bohrium
Bohrium AI for Science platform is dedicated to providing a new research paradigm for scientists worldwide in the AI era, empowering scientific exploration through technological innovation and making the acquisition, understanding, and application of knowledge more intelligent and efficient. OrigeneMCP can be directly accessed and invoked through the Bohrium AI for Science platform.
4. Cite OrigeneMCP
Any publication that discloses findings arising from using this source code should cite: